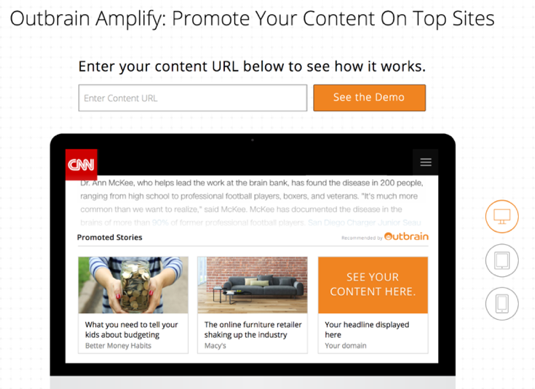
|
|
頭條 |
| H1 |
H2 |
H3 |
H4 |
H5 |
H6 |
| 1 |
5 |
0 |
0 |
0 |
0 |
- [H1] New and Noteworthy
- [H2] About AspGD
- [H2] Upcoming Meetings & Courses
- [H2] Server maintenance tomorrow night
- [H2] Title of item
text of item here
(Posted DATE, 2012)
-->
Aspergillus annotation updates
The reference gene sets in AspGD have been updated, based on PASA analysis conducted by AspGD in 2015. The following changes were made:
A. nidulans - 5 new features were added, and 4 former pseudogenes were reannotated as uncharachterized ORFs;
A. fumigatus - 2,473 features were affected: 59 new features were added; the sequences for 242 features were extended and 154 shortened; 4 features were merged to create 2 new features; 1,757 5-prime UTRs and 1,333 3-prime UTRs were added or modified; terminal exons were added to 313 features; the intron/exon structure was modified for 389 features;
A. niger - two new features were added (An11g02363 and An17g02286);
A. oryzae - 7,279 features were affected: 287 new features were added; the sequences for 706 features were extended and 192 shortened; 192 features were merged to create 93 new feature; 5,141 5-prime UTRs and 5,213 3-prime UTRs were added or modified; terminal exons were added to 897 features; the intron/exon structure was modified for 500 features.
Please, visit the Chromosome History page to review the changes.
(Posted September 15, 2015)
AspGD Curation News
- [H2] AspGD and FungiDB integration
AspGD data are being integrated into FungiDB. Please click here for additional details.
Discussion of how to maximize the value of FungiDB for the Aspergillus research community will be a major topic at the upcoming
AsperFest12 meeting at Asilomar (March 16-17, 2015).
An open letter to the Aspergillus research community on genome database resources:
As many of you may already be aware, the NIH R01 research grant that has supported development of the AspGD database will not be
renewed … not because the NIH doesn't appreciate the value and importance of this resource to the Aspergillus research community,
but because the current funding climate makes it difficult to support curation-intensive databases serving individual pathogens
and relatively small research communities. However, NIH has provided one year of funding to ensure that information from AspGD
is integrated into FungiDB, a component of the EuPathDB Bioinformatics Resource Center. We do not anticipate that AspGD will
disappear: you should still be able to access the existing web site, at least for the foreseeable future, but curatorial
engagement will have to be significantly reduced over the coming months.
The EuPathDB family of databases supports a wide range of microbial eukaryotes, including protozoan parasites and many fungal and
oomycete species, including non-pathogens. This resource has been specifically designed to provide sustainable, cost-effective
automated analysis of multiple genomes, integrating curated information where available, along with comments and supporting
evidence from the user community (PubMedIDs, phenotypic information, images, datasets, etc). In addition to gene records,
browser views, and data downloads, FungiDB also provides a variety of sophisticated tools for integrating and mining diverse
Omics datasets, which we expect that Aspergillus researchers and other fungal biologists will find quite useful. See sidebar on
the FungiDB web site for access to tutorials, videos, and exercises; more fungi-specific materials should become available over
the coming months.
AspGD and FungiDB/EuPathDB will be working together to maximize value to the Aspergillus community, given the limited resources
available. The upcoming AsperFest12 and 28th Fungal Genetics meetings in Asilomar (March 16-22, 2015) will be attended by staff
from both groups, with presentations and a ‘help-desk’ to introduce the Aspergillus community to FungiDB. We look forward to
productive discussions on how to ensure that Aspergillus researchers retain access to information from AspGD, learn to use
FungiDB effectively and have a voice in its further development, and craft a strategy for meeting future informatics needs.
Jessica Kissinger (Univ Georgia) - Joint-PI, EuPathDB
Michelle Momany (Univ Georgia) - Community Liaison, AspGD & FungiDB
David Roos (Univ Pennsylvania) - Joint-PI, EuPathDB; Co-PI, FungiDB
Gavin Sherlock (Stanford Univ) - Principal Investigator, AspGD
Jason Stajich (UC Riverside) - Principal Investigator, FungiDB
Jennifer Wortman (Broad Inst) - Co-PI, AspGD
(Posted February, 2015)
Sequence trace files available
We have added archival Aspergillus sequence data from the NCBI TraceDB site, and these data are now available
from our AspGD download site.
(Posted November 8, 2013)
New "Big 4" ortholog alignment
We have added a new, streamlined, four-species protein alignment to the ortholog cluster pages in our Sybil Comparative Genomics
Viewer. This "Big 4" alignment displays a Clustal protein alignment of the orthologs from A. nidulans FGSC A4, A.
oryzae RIB40, A. fumigatus Af293 and A. niger CBS513.88 (see example). As we continue to add new species into our Sybil
Comparative Genomics Viewer, the number of proteins represented in each multispecies ortholog cluster has grown significantly. We
continue to provide an alignment of all of the orthologs from the full set of species, and now include the "Big 4" alignment in
addition. We hope this feature is helpful, and we welcome your feedback any time.
(Posted August 13, 2013)
A. niger annotation updates
The A. niger reference gene set has been updated, based on PASA analysis. 4,123 features were affected: 543 CDSs were
extended and 195 shortened; 30 features were merged into 15 features; 2,907 5-prime UTRs and 2,601 3-prime UTRs were added or
modified; New exons were added to the ends of 270 features, and the internal intron/exon structure was modified for 491 features.
The GBrowse Genome Browser displays the old annotation as a "Historic Track" to facilitate visual comparison, for example, of new UTRs or gene model structure updates, for example, a gene merge. The current and historic reference annotation can be viewed alongside RNA-Seq transcript
data in AspGD JBrowse (zoom in to see spliced reads across intronic regions, including the intron within what was previously
annotated as an intergenic region).
(Posted August 13, 2013)
Introducing JBrowse for AspGD
JBrowse is now available for exploring large-scale data in AspGD, including RNA-Seq data for A. nidulans FGSC A4, RNA-Seq and
genomic variation data for A. fumigatus Af293, and RNA-Seq data for A. niger CBS 513.88 and A. oryzae RIB40. To see
the large-scale data in the region of any gene, select the JBrowse link on the Locus Summary page, for example, in the "RNA-Seq
Data" section of the A. nidulans veA page. The JBrowse software has been developed as part of the GMOD project. The links to the GenomeView browser are no longer displayed
on the Locus Summary pages due to incompatibilities of the browser software software with some web browsers and versions of Java.
Genome View may still be accessed from the Search Options page. (Posted June 19,
2013)
RNA-Seq data used to improve reference annotation for A. fumigatus
We have made significant updates to the structural annotation of the Aspergillus fumigatus Af293 reference genome based on
PASA analysis using RNA-Seq data (from Müller et al. [2012]) to provide experimental support for gene
model modifications. The coordinates of 5,174 features were updated: 1,044 CDSs were extended and 641 shortened; 28 features
were merged into 14 features; 4,524 5-prime UTRs and 3,545 3-prime UTRs were added or modified; New exons were added to the ends
of 1,255 features, and the internal intron/exon structure was modified for 1,133 features.
In addition, we have updated the structural annotation of the Aspergillus fumigatus A1163 (CEA10) strain, which is not the
reference strain, but for which we provide sequence files, protein domain files, and mulispecies BLAST capabilities. The PASA analysis of A. fumigatus A1163 utilized RNA-Seq data
provided by JCVI (SRA project SRP003796).
(Posted April 2, 2013)
Archived News
HEADING段落標題也許是除了網頁標題TITLE之外,搜索引擎要搜索的信息,
它告訴搜索引擎關於你文章段落的大概內容,建議撰寫 HEADING,從 H1-H6 到你的網頁裏面。
 使用在標題區域的關鍵詞研究方法
使用在標題區域的關鍵詞研究方法
|

 標題優化:
標題優化: 怎樣在你的網頁加入SEO標題標簽?
怎樣在你的網頁加入SEO標題標簽?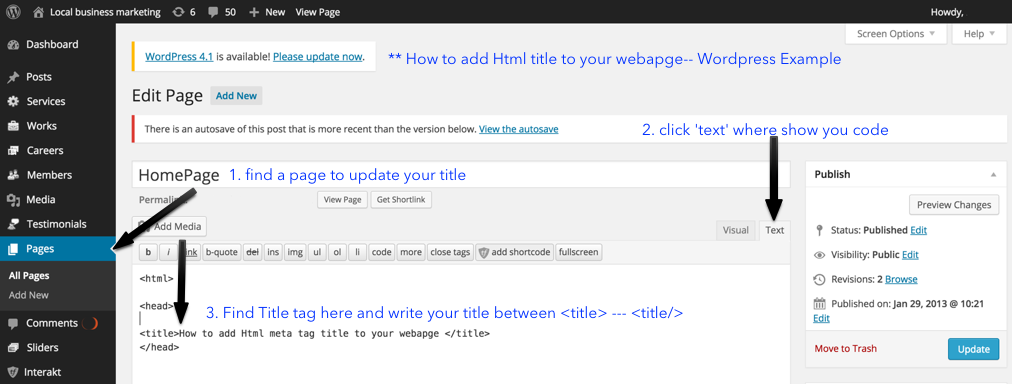
 怎樣搜索盈利性的關鍵詞標題?
怎樣搜索盈利性的關鍵詞標題?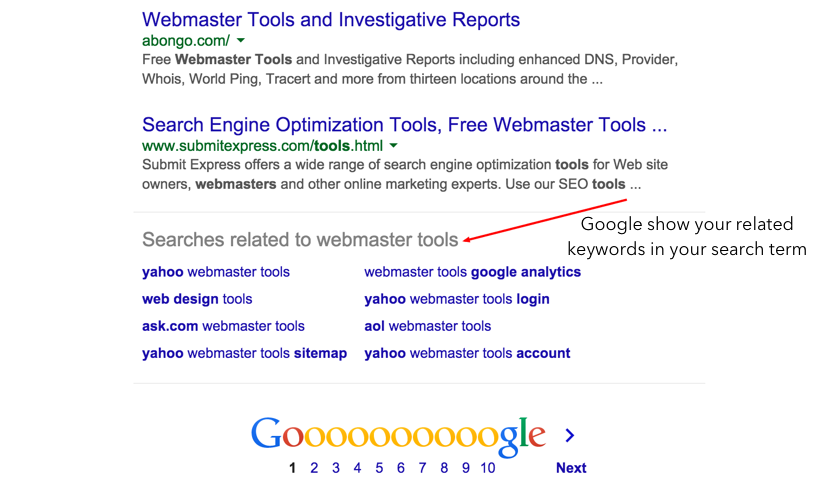
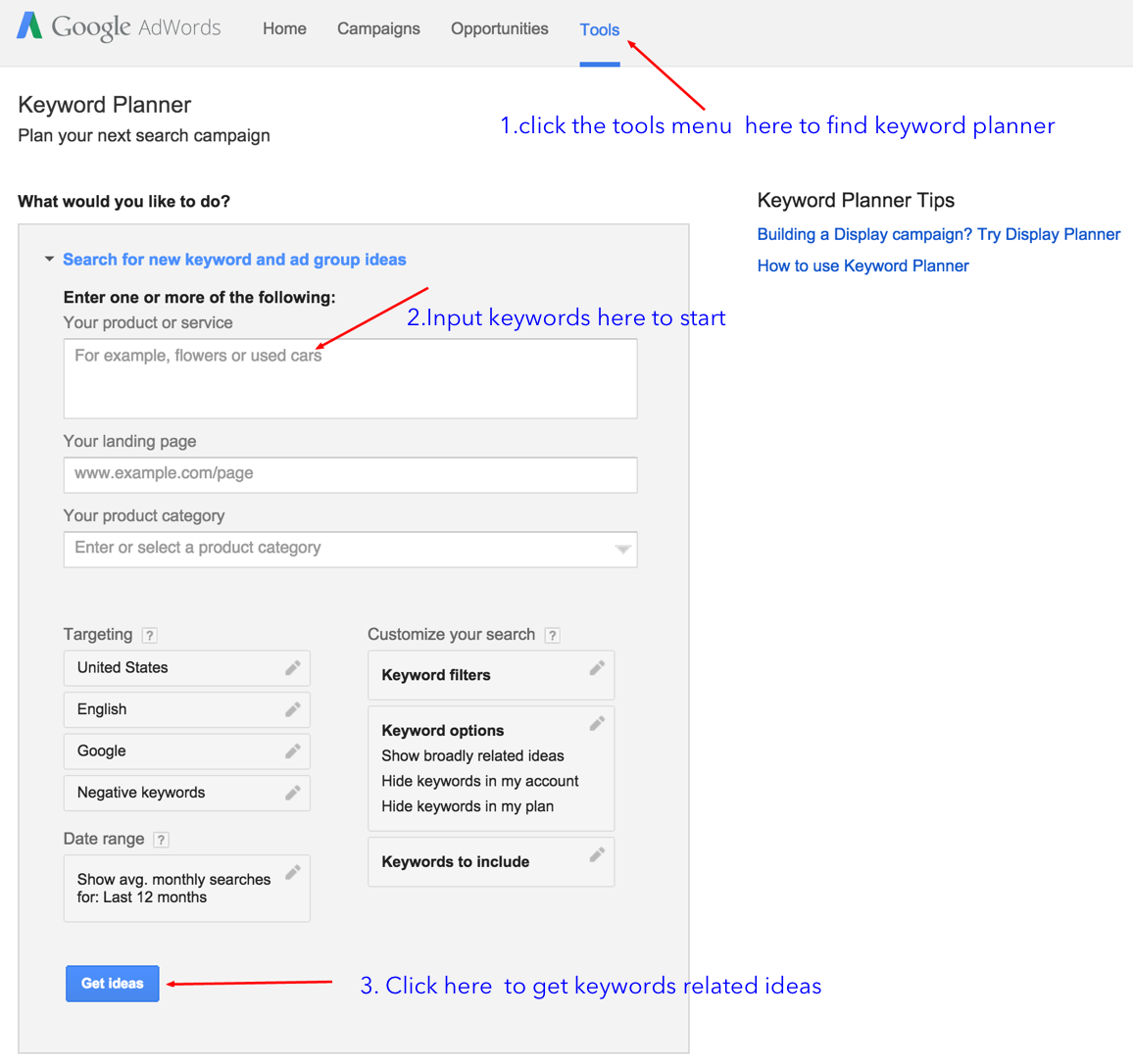
 關鍵詞詞組建議工具
關鍵詞詞組建議工具
 預覽網頁標題源代碼
預覽網頁標題源代碼 預覽標題源代碼工具
預覽標題源代碼工具
 了解谷歌搜索工作原理?
了解谷歌搜索工作原理? 了解谷歌搜索工作原理?
了解谷歌搜索工作原理?





 過關,可以休息了
過關,可以休息了 還可以,建議繼續優化
還可以,建議繼續優化 警告,建議修復
警告,建議修復 錯誤,需要修復
錯誤,需要修復
 SEO內容
SEO內容


 SEO鏈接
SEO鏈接
 SEO 關鍵詞
SEO 關鍵詞


 可用性建議
可用性建議

 網頁文本
網頁文本
 移動設備
移動設備

 分析工具
分析工具
 營銷策略
營銷策略
 网站各类排名
网站各类排名



 关键词排名
关键词排名
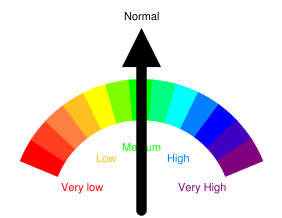 关键词难度工具
关键词难度工具
 网站安全
网站安全

 站长推荐工具
站长推荐工具